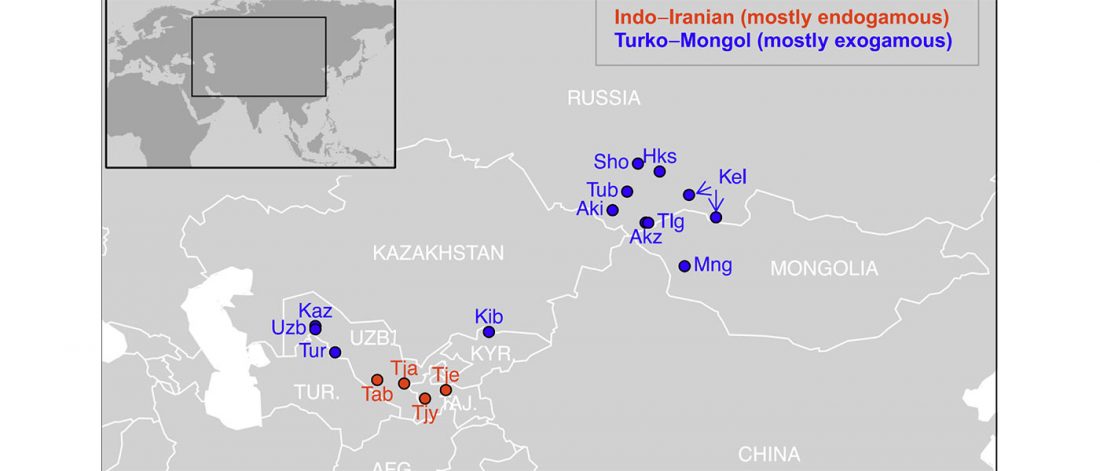
Close inbreeding and low genetic diversity in Inner Asian human populations despite geographical exogamy
Open access Close inbreeding and low genetic diversity in Inner Asian human populations despite geographical exogamy, by Marchi et al. Scientific Reports (2018) 8:9397.
Abstract (emphasis mine):
… Read the rest “Close inbreeding and low genetic diversity in Inner Asian human populations despite geographical exogamy”When closely related individuals mate, they produce inbred offspring, which often have lower fitness than outbred ones. Geographical exogamy, by favouring matings between distant individuals, is thought to be an inbreeding avoidance mechanism; however, no data has clearly tested this prediction. Here, we took advantage of the diversity of matrimonial systems in humans to explore the impact of geographical exogamy on genetic diversity and inbreeding. We collected ethno-demographic data for 1,344 individuals