RISE1.SG, R1b from Poland CWC, a likely mislabelled Balto-Slav
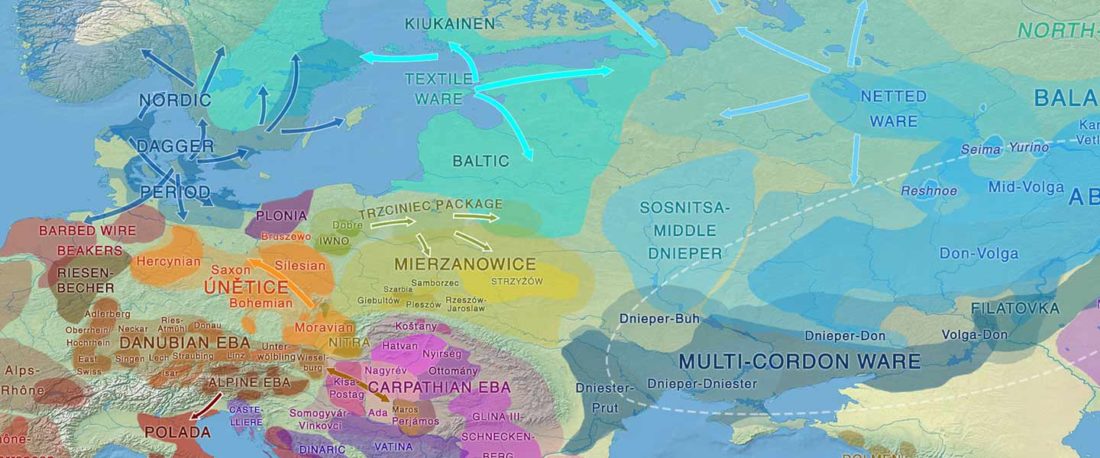
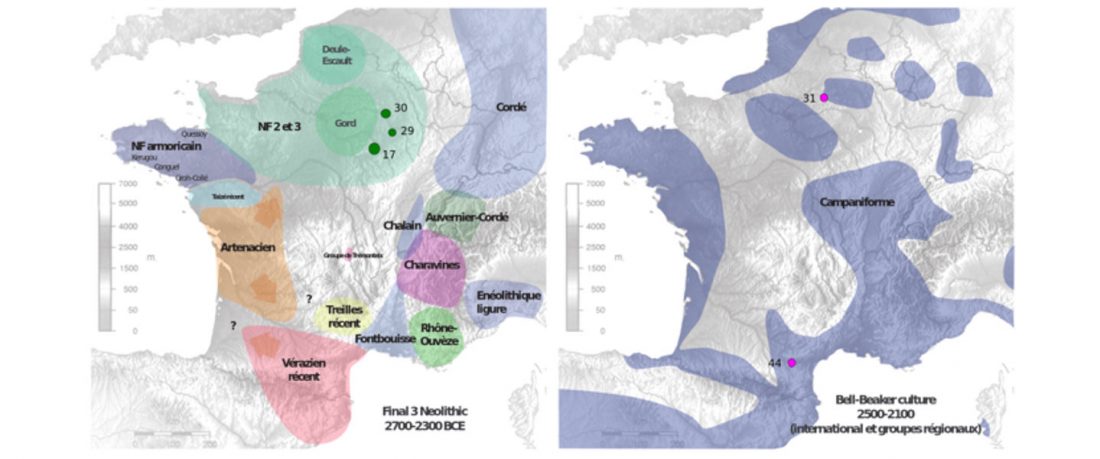
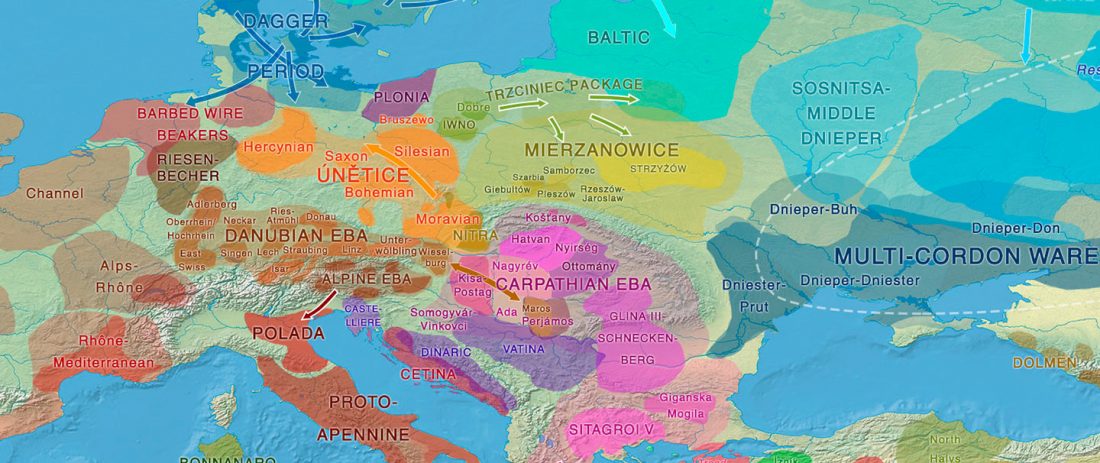
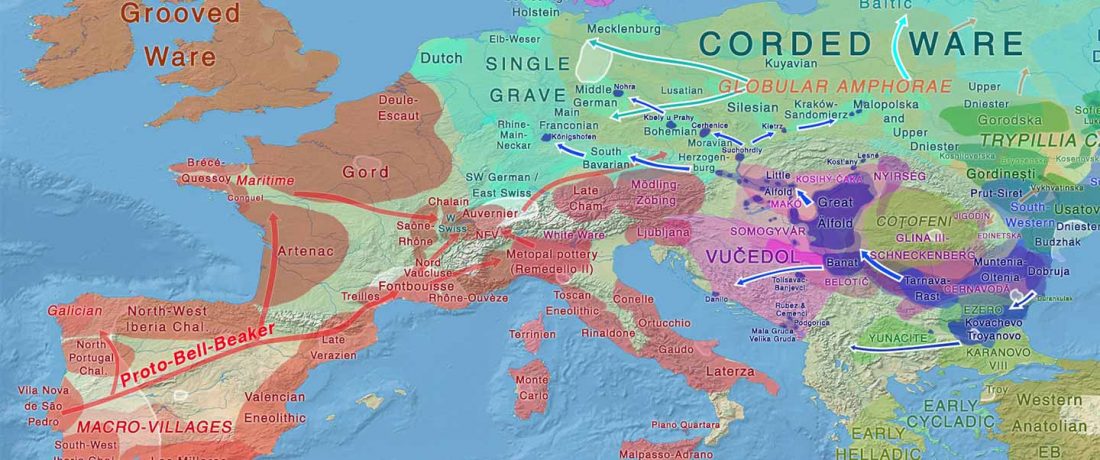
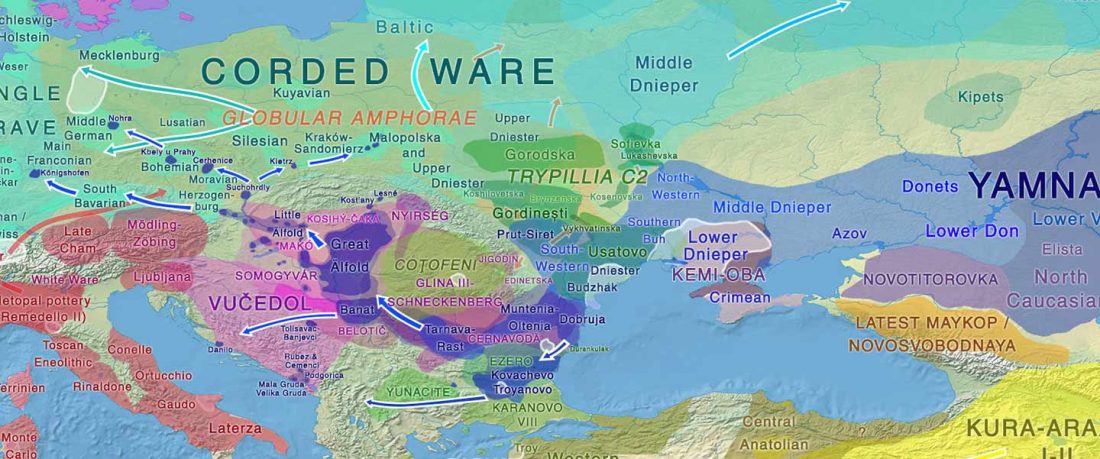
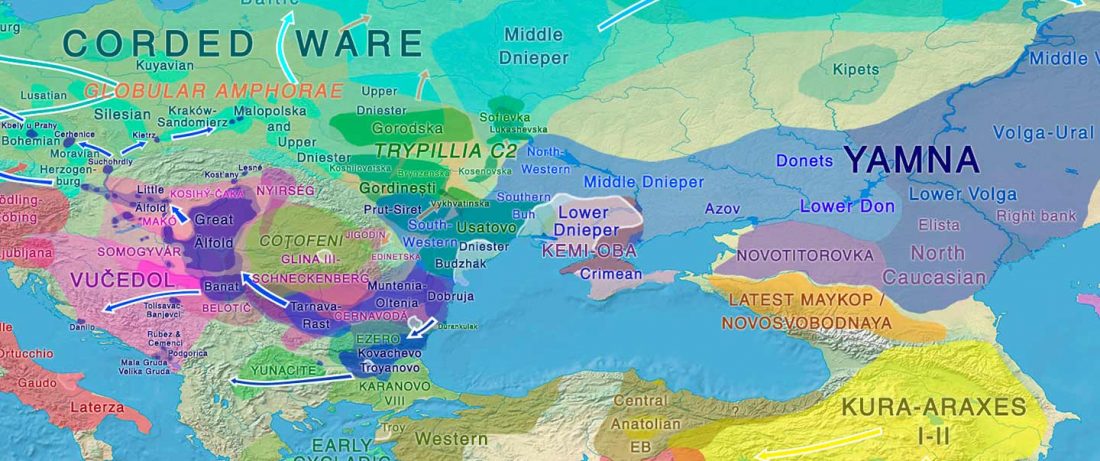
When I started ruminating in 2016 over the apparent differences between populations that kept the two-velar distinction of Indo-Tocharian, and the only two unrelated dialectal groups that showed a strong satemization trend, I believed that – much like in modern times – there would be no clear-cut division in terms of ancestry or Y-DNA haplogroups between neighbouring forest-steppe and steppe populations.
The answer to the question of interacting ethnolinguistic groups had to lie, as everything else, on the investigation of fine-scale population movements that must have put Uralic-speaking peoples as the main substratum of Balto-Slavic and Indo-Iranian.
Still, … Read the rest “RISE1.SG, R1b from Poland CWC, a likely mislabelled Balto-Slav”