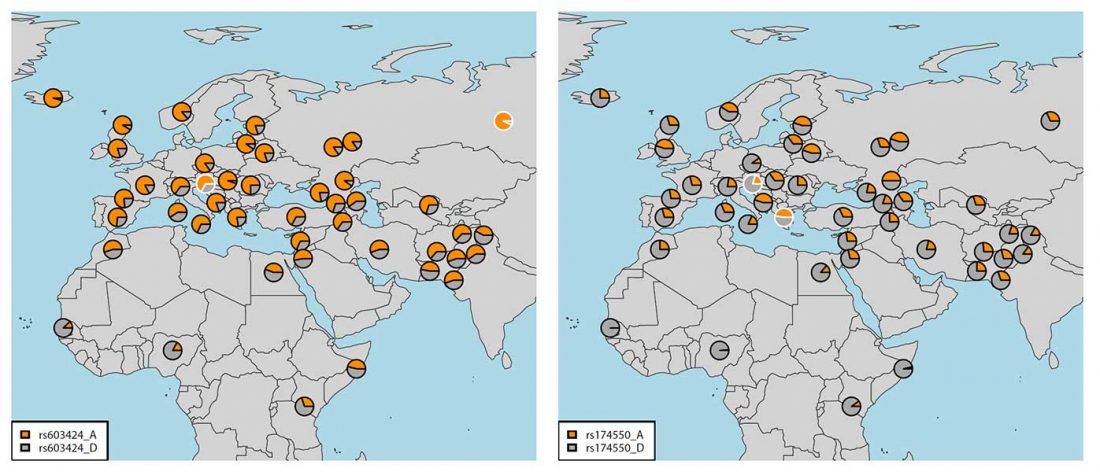
Open access Genetic Landscape of Slovenians: Past Admixture and Natural Selection Pattern, by Maisano Delser et al. Front. Genet. (2018).
Interesting excerpts (emphasis mine):
Samples
Overall, 96 samples ranging from Slovenian littoral to Lower Styria were genotyped for 713,599 markers using the OmniExpress 24-V1 BeadChips (Figure 1), genetic data were obtained from Esko et al. (2013). After removing related individuals, 92 samples were left. The Slovenian dataset has been subsequently merged with the Human Origin dataset (Lazaridis et al., 2016) for a total of 2163 individuals.
Y chromosome
… Read the rest “Genetic landscape and past admixture of modern Slovenians”First, Y chromosome genetic diversity was assessed. A total of