New paper (behind paywall), Ancient genomes from Iceland reveal the making of a human population, by Ebenesersdóttir et al. Science (2018) 360(6392):1028-1032.
Abstract and relevant excerpts (emphasis mine):
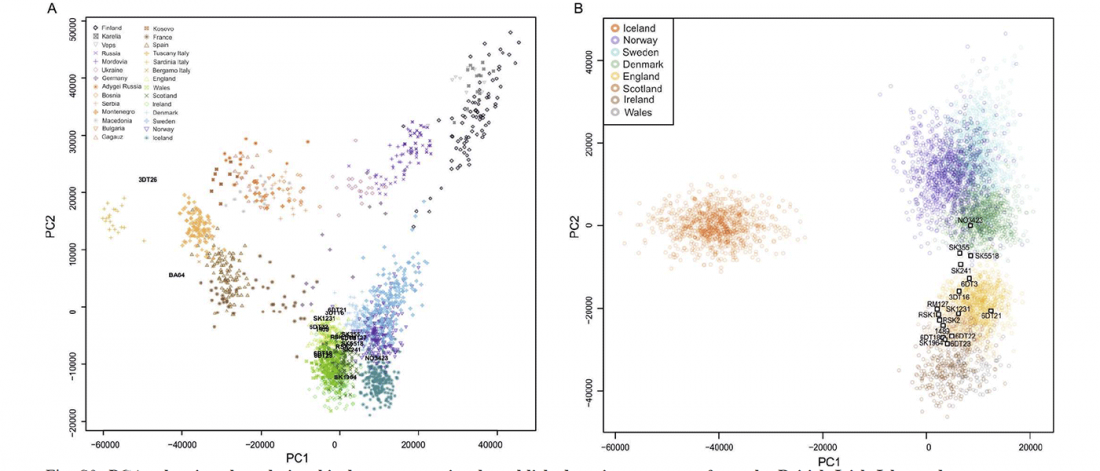
… Read the rest “Reproductive success among ancient Icelanders stratified by ancestry”Opportunities to directly study the founding of a human population and its subsequent evolutionary history are rare. Using genome sequence data from 27 ancient Icelanders, we demonstrate that they are a combination of Norse, Gaelic, and admixed individuals. We further show that these ancient Icelanders are markedly more similar to their source populations in Scandinavia and the British-Irish Isles than to contemporary Icelanders, who have been shaped by 1100 years of