New paper (behind paywall) Sahelian pastoralism from the perspective of variants associated with lactase persistence, by Priehodová et al. Am J Phys Anthropol (2020) e24116.
Interesting excerpts from the discussion (emphasis mine, minor modifications for clarity):
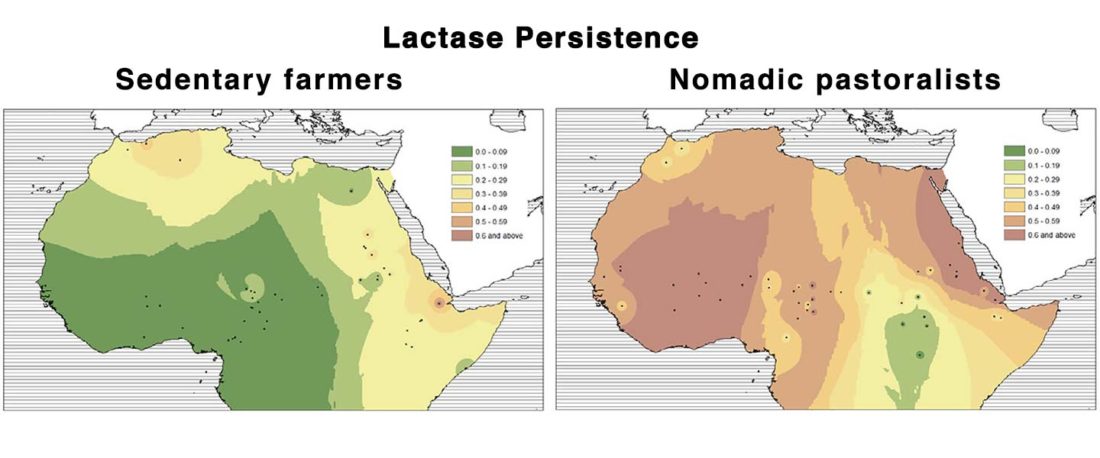
… Read the rest “Neolithic spread of “Eurasian” Lactase Persistence among Saharan pastoralists”Our investigation of LP variant frequencies revealed new and interesting results related to the origins of pastoralism and subsequent gene flow between pastoralists and farmers in the Sahel/Savannah belt of Africa.
- We observed a clear distinction between regions west and east of Lake Chad: while variant −13910*T prevails in the western Sahel, where we found it only in pastoralists such as the Fulani,