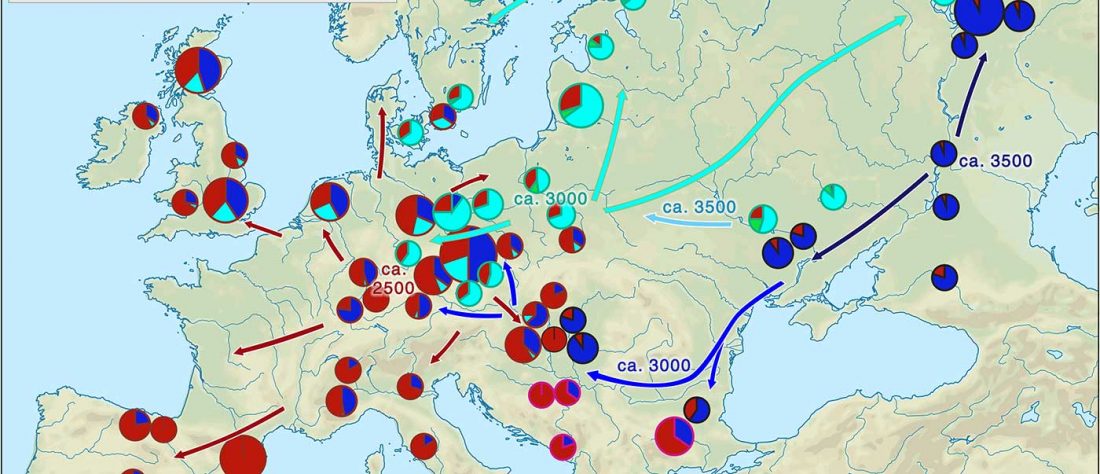
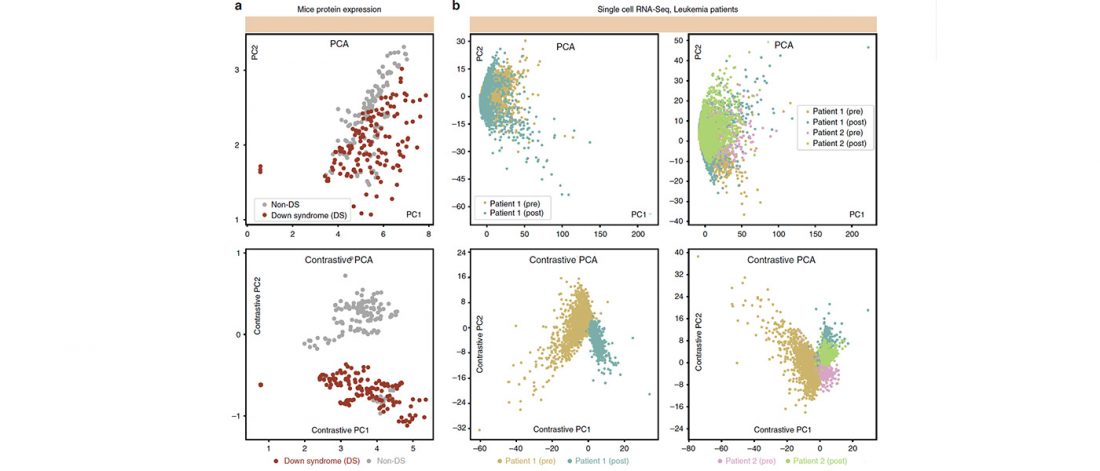
The title says it all. I have used some free time to update the series A Song of Sheep and Horses:
- Book One: A Song of Sheep and Horses: eurafrasia nostratica, eurasia indouralica. Languages.
- Books Two & Three: A Game of Clans: collectores venatoresque, agricolae pastoresque & A Clash of Chiefs: rex militaris, rex sacrorum. Cultures and peoples.
- Book Four: A Storm of Hordes: hic sunt leones, hic sunt dracones. Maps and graphics.
I basically added information from the latest papers published, which (luckily enough for me) haven’t been too … Read the rest “Updates to ASoSaH: new maps, updated PCA, and added newest research papers”