New and interesting research these days in BioRxiv:
Haplotype sharing provides insights into fine-scale population history and disease in Finland, by Martín et al. (2017):
Finland provides unique opportunities to investigate population and medical genomics because of its adoption of unified national electronic health records, detailed historical and birth records, and serial population bottlenecks. We assemble a comprehensive view of recent population history (≤100 generations), the timespan during which most rare disease-causing alleles arose, by comparing pairwise haplotype sharing from 43,254 Finns to geographically and linguistically adjacent countries with different population histories, including 16,060 Swedes, Estonians, Russians, and Hungarians. We find much more extensive sharing in Finns, with at least one ≥ 5 cM tract on average between pairs of unrelated individuals. By coupling haplotype sharing with fine-scale birth records from over 25,000 individuals, we find that while haplotype sharing broadly decays with geographical distance, there are pockets of excess haplotype sharing; individuals from northeast Finland share several-fold more of their genome in identity-by-descent (IBD) segments than individuals from southwest regions containing the major cities of Helsinki and Turku. We estimate recent effective population size changes over time across regions of Finland and find significant differences between the Early and Late Settlement Regions as expected; however, our results indicate more continuous gene flow than previously indicated as Finns migrated towards the northernmost Lapland region. Lastly, we show that haplotype sharing is locally enriched among pairs of individuals sharing rare alleles by an order of magnitude, especially among pairs sharing rare disease causing variants. Our work provides a general framework for using haplotype sharing to reconstruct an integrative view of recent population history and gain insight into the evolutionary origins of rare variants contributing to disease.
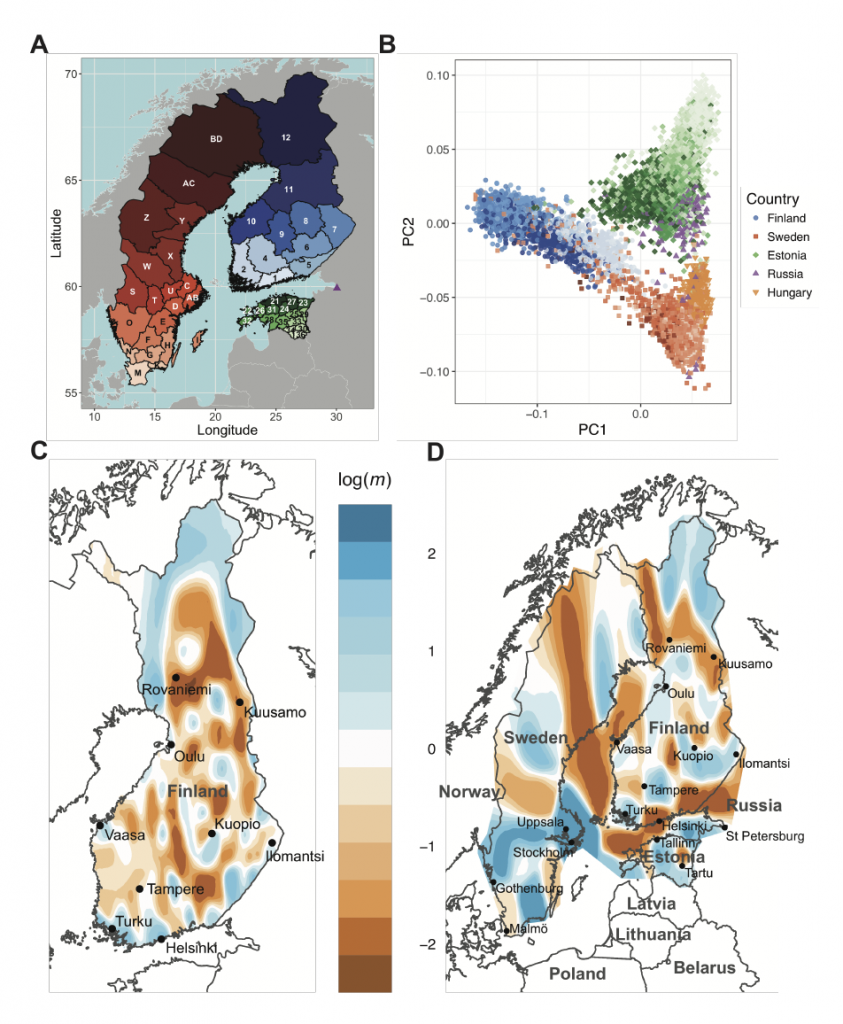
Interesting to understand this paper is the whole research published by the Institute for Molecular Medicine Finland (FIMM): their website contains detailed research on Finland’s recent genetic history.
NOTE: The featured image of this article contains three figures from the FIMM (License CC-BY 4.0). Left: Position of the points represents the locations of 1042 Finnish individuals. By clustering the individuals into two groups based on genome data we see a split between eastern (blue) and western (red) parts. Individuals who show considerable relatedness to both groups have been colored with cyan. Both parents of each individual were born close to each other and based on the parents’ birth years we can infer that we are looking at the genetic structure present in Finland before 1950s. Center: An estimated borderline of the Treaty of Nöteborg on top of the map from the left. The border line is drawn between Jääski (28.92 N, 61.04 E) and Pyhäjoki (24.26 N, 64.46 E). Right: The settlement border divides Finland into the early settlement region (to west and south of the border) and the late settlement region (to east and north of the border) (Jutikkala 1933, s. 91). We see that Southern Savo (in south-eastern part of the early settlement) is among the only parts of the early settlement region that is dominated by the eastern genetic group. Information from Matti Pirinen and Sini Kerminen, 24.5.2017.
An Unexpectedly Complex Architecture for Skin Pigmentation in Africans, by Martin et al (2017):
Fewer than 15 genes have been directly associated with skin pigmentation variation in humans, leading to its characterization as a relatively simple trait. However, by assembling a global survey of quantitative skin pigmentation phenotypes, we demonstrate that pigmentation is more complex than previously assumed with genetic architecture varying by latitude. We investigate polygenicity in the Khoe and the San, populations indigenous to southern Africa, who have considerably lighter skin than equatorial Africans. We demonstrate that skin pigmentation is highly heritable, but that known pigmentation loci explain only a small fraction of the variance. Rather, baseline skin pigmentation is a complex, polygenic trait in the KhoeSan. Despite this, we identify canonical and non-canonical skin pigmentation loci, including near SLC24A5, TYRP1, SMARCA2/VLDLR, and SNX13 using a genome-wide association approach complemented by targeted resequencing. By considering diverse, under-studied African populations, we show how the architecture of skin pigmentation can vary across humans subject to different local evolutionary pressures.
Contrasting maternal and paternal genetic variation of hunter-gatherer groups in Thailand, by Kutanan et al. (2017):
The Maniq and Mlabri are the only recorded nomadic hunter-gatherer groups in Thailand. Here, we sequenced complete mitochondrial (mt) DNA genomes and ~2.364 Mbp of non-recombining Y chromosome (NRY) to learn more about the origins of these two enigmatic populations. Both groups exhibited low genetic diversity compared to other Thai populations, and contrasting patterns of mtDNA and NRY diversity: there was greater mtDNA diversity in the Maniq than in the Mlabri, while the converse was true for the NRY. We found basal uniparental lineages in the Maniq, namely mtDNA haplogroups M21a, R21 and M17a, and NRY haplogroup K. Overall, the Maniq are genetically similar to other negrito groups in Southeast Asia. By contrast, the Mlabri haplogroups (B5a1b1 for mtDNA and O1b1a1a1b and O1b1a1a1b1a1 for the NRY) are common lineages in Southeast Asian non-negrito groups, and overall the Mlabri are genetically similar to their linguistic relatives (Htin and Khmu) and other groups from northeastern Thailand. In agreement with previous studies of the Mlabri, our results indicate that the Malbri do not directly descend from the indigenous negritos. Instead, they likely have a recent origin (within the past 1,000 years) by an extreme founder event (involving just one maternal and two paternal lineages) from an agricultural group, most likely the Htin or a closely-related group.
Related:
- Another hint at the role of Corded Ware peoples in spreading Uralic languages into north-eastern Europe, found in mtDNA analysis of the Finnish population
- Wiik’s theory about the spread of Uralic into east and central Europe, and the Uralic substrate in Germanic and Balto-Slavic
- New Ukraine Eneolithic sample from late Sredni Stog, near homeland of the Corded Ware culture
- Germanic–Balto-Slavic and Satem (‘Indo-Slavonic’) dialect revisionism by amateur geneticists, or why R1a lineages *must* have spoken Proto-Indo-European
- Something is very wrong with models based on the so-called ‘steppe admixture’ – and archaeologists are catching up