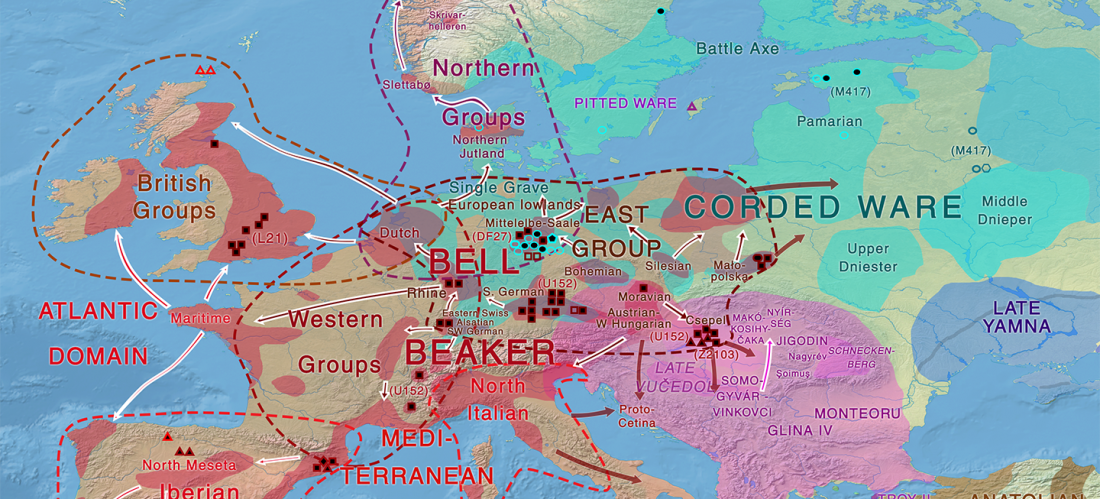
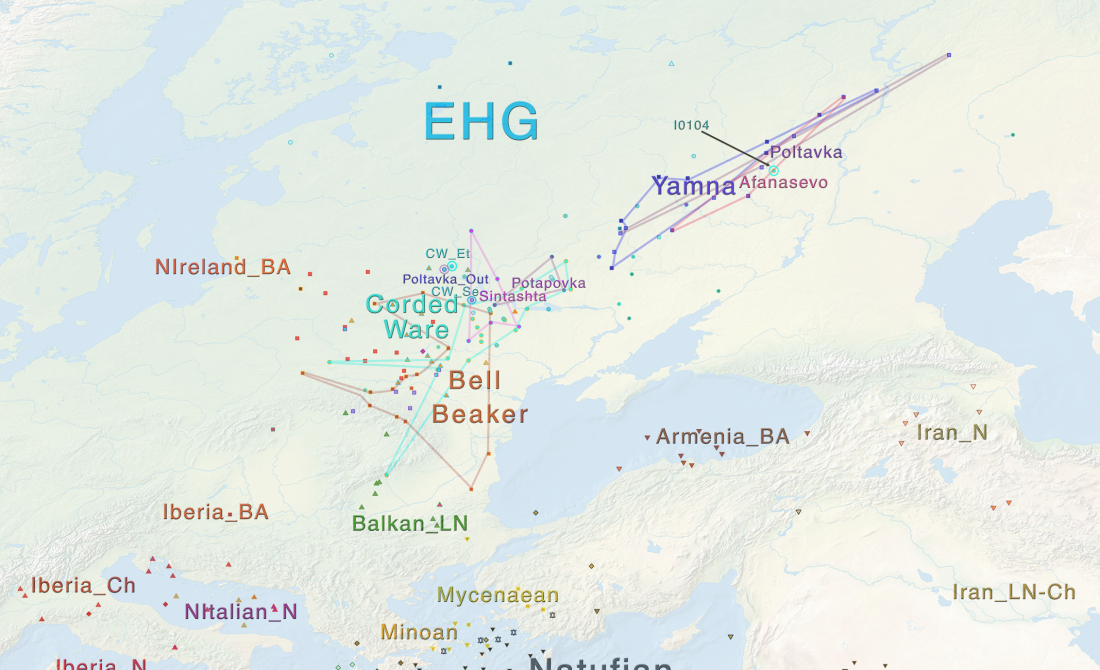
Open access Estimating genetic kin relationships in prehistoric populations, by Monroy Kuhn, Jakobsson, & Günther, PLOS One (2018).
Abstract
… Read the rest “Estimating genetic kin relationships in prehistoric populations: the Corded Ware family from Esperstedt”Archaeogenomic research has proven to be a valuable tool to trace migrations of historic and prehistoric individuals and groups, whereas relationships within a group or burial site have not been investigated to a large extent. Knowing the genetic kinship of historic and prehistoric individuals would give important insights into social structures of ancient and historic cultures. Most archaeogenetic research concerning kinship has been restricted to uniparental markers, while studies using genome-wide information were mainly focused on comparisons between populations. Applications