Neanderthal language revisited: not only us, by Dediu and Levinson, Curr Opin Behav Sci (2018) 21:49–55.
Abstract:
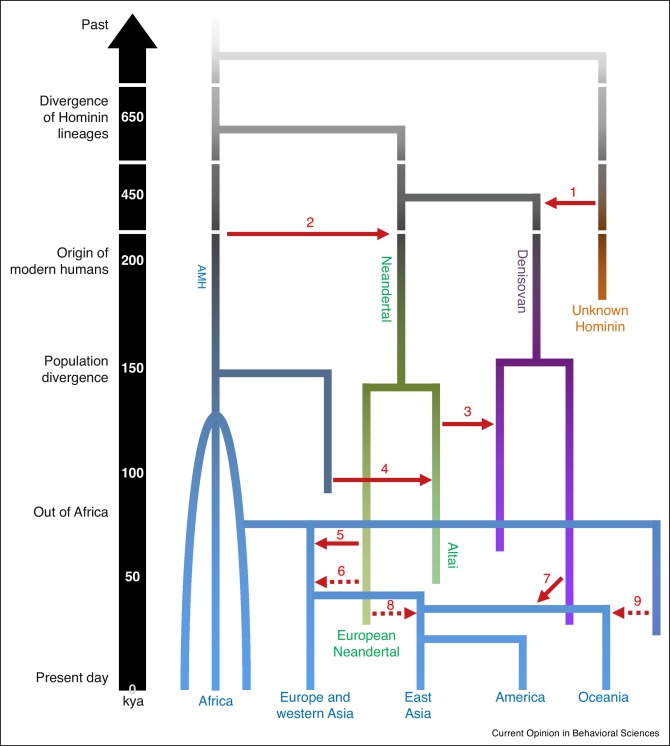
Here we re-evaluate our 2013 paper on the antiquity of language (Dediu and Levinson, 2013) in the light of a surge of new information on human evolution in the last half million years. Although new genetic data suggest the existence of some cognitive differences between Neanderthals and modern humans — fully expected after hundreds of thousands of years of partially separate evolution, overall our claims that Neanderthals were fully articulate beings and that language evolution was gradual are further substantiated by the wealth of new genetic, paleontological and archeological evidence briefly reviewed here.
Check out also the interesting open access article Drawings of Representational Images by Upper Paleolithic Humans and their Absence in Neanderthals Might Reflect Historical Differences in Hunting Wary Game, by Richard G. Coss.
Related:
- Language evolution and language change related to ancient DNA
- On the origin of language and human evolution
- The myth of mixed language, the concepts of culture core and package, and the invention of ‘Steppe folk’
- Evolutionary forces in language change depend on selective pressure, but also on random chance
- Forces driving grammatical change are different to those driving lexical change