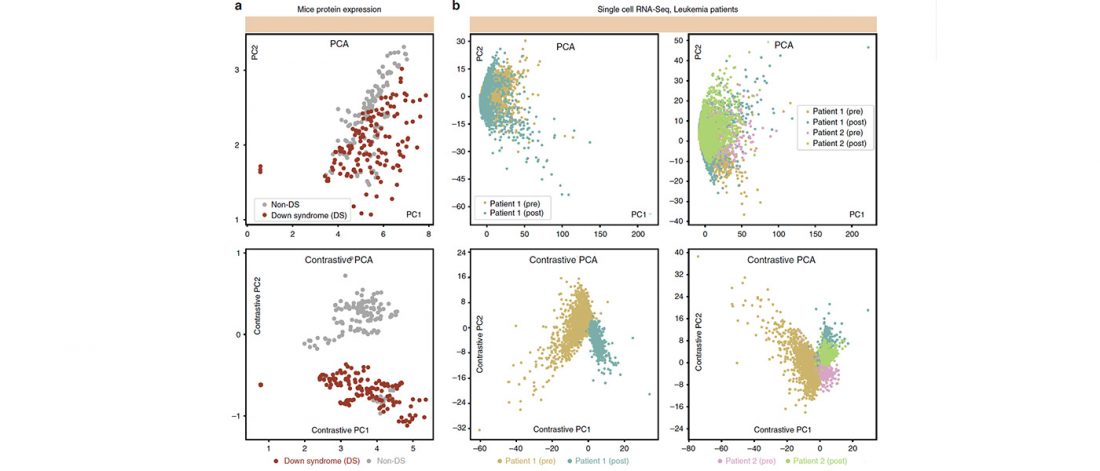
Interesting open access paper Exploring patterns enriched in a dataset with contrastive principal component analysis, by Abid, Zhang, Bagaria & Zou, Nature Communications (2018) 9:2134.
Abstract (emphasis mine):
… Read the rest “Contrastive principal component analysis (cPCA) to explore patterns specific to a dataset”Visualization and exploration of high-dimensional data is a ubiquitous challenge across disciplines. Widely used techniques such as principal component analysis (PCA) aim to identify dominant trends in one dataset. However, in many settings we have datasets collected under different conditions, e.g., a treatment and a control experiment, and we are interested in visualizing and exploring patterns that are specific to one dataset. This paper proposes a method, contrastive principal component analysis