Open access preprint Cultural Innovations influence patterns of genetic diversity in Northwestern Amazonia, by Arias et al., bioRxiv (2018).
Abstract (emphasis mine):
Human populations often exhibit contrasting patterns of genetic diversity in the mtDNA and the non-recombining portion of the Y-chromosome (NRY), which reflect sex-specific cultural behaviors and population histories. Here, we sequenced 2.3 Mb of the NRY from 284 individuals representing more than 30 Native-American groups from Northwestern Amazonia (NWA) and compared these data to previously generated mtDNA genomes from the same groups, to investigate the impact of cultural practices on genetic diversity and gain new insights about NWA population history. Relevant cultural practices in NWA include postmarital residential rules and linguistic-exogamy, a marital practice in which men are required to marry women speaking a different language. We identified 2,969 SNPs in the NRY sequences; only 925 SNPs were previously described. The NRY and mtDNA data showed that males and females experienced different demographic histories: the female effective population size has been larger than that of males through time, and both markers show an increase in lineage diversification beginning ~5,000 years ago, with a male-specific expansion occurring ~3,500 years ago. These dates are too recent to be associated with agriculture, therefore we propose that they reflect technological innovations and the expansion of regional trade networks documented in the archaeological evidence. Furthermore, our study provides evidence of the impact of postmarital residence rules and linguistic exogamy on genetic diversity patterns. Finally, we highlight the importance of analyzing high-resolution mtDNA and NRY sequences to reconstruct demographic history, since this can differ considerably between males and females.
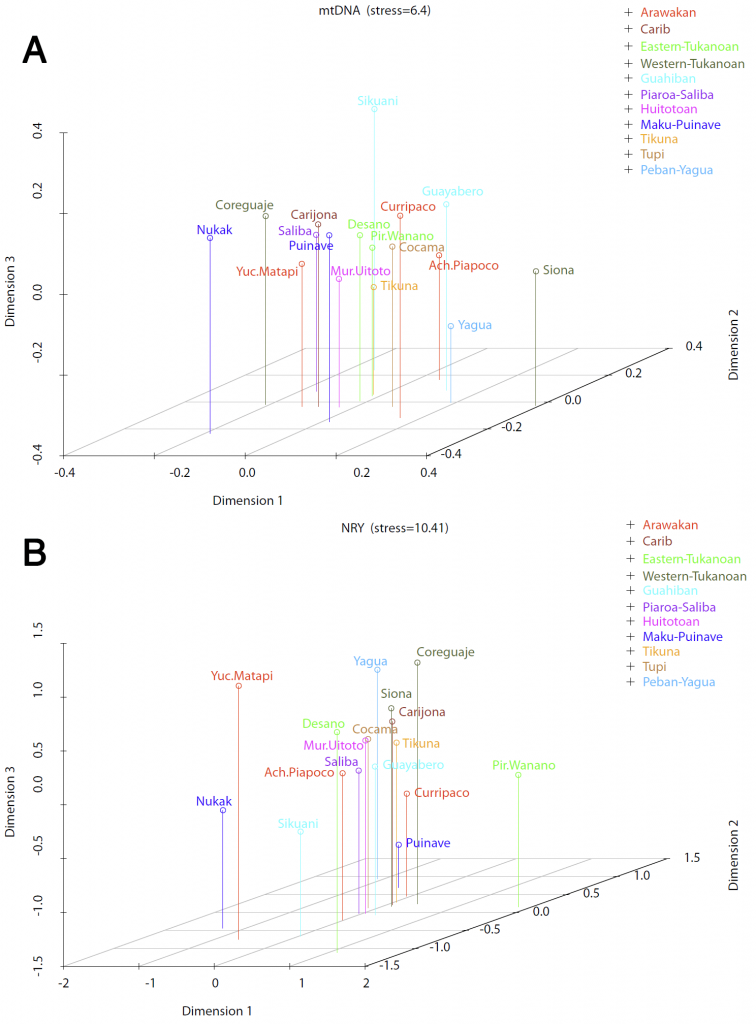
Looking more precisely at the different groups (even with the resampling approach), there are no significant differences between matrilocal and patrilocal groups. At best, as the study proposes, “this is just one of the factors at play in structuring the observed genetic variation”.
Interesting excerpts:
(…) we found evidence that the patterns of genetic differentiation depend on the geographical scale of the study. The magnitude of between-population differentiation in the NRY compared to the mtDNA is smaller when looking at the continental scale than in NWA (Figure 6). This is in agreement with the findings of Wilkins and Marlowe (2006), who showed that the excess of between-population differentiation for the NRY in comparison to the mtDNA decreases when comparing more geographically distant populations. Heyer et al. (2012) and Wilkins and Marlowe (2006) have proposed that at a local scale the patterns of genetic diversity reflect cultural practices over a relatively small number of generations, whereas at a larger geographic scale the genetic diversity reflects old migration and/or old common ancestry patterns(Heyer et al. 2012; Wilkins and Marlowe 2006).
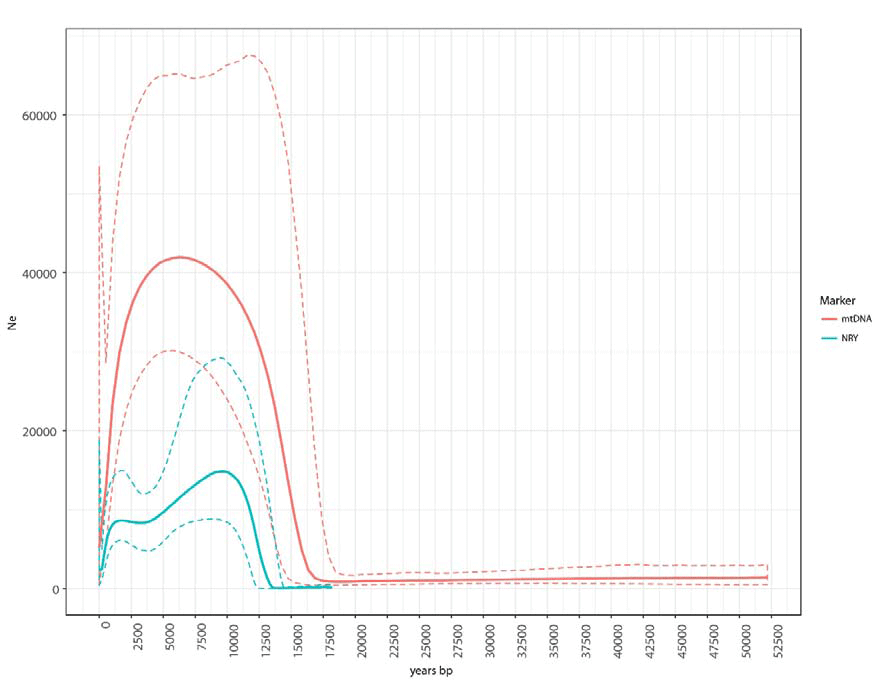
The BSP plots and the diversity statistics indicate that overall the Ne of males has been smaller than that of females. One tentative explanation for this difference is that it reflects larger differences in reproductive success among males than among females. Some support for this explanation comes from the shape of the phylogenies (Supplementary Figures 1 and 6), since differences in reproductive success and the cultural transmission of fertility lead to imbalance phylogenies (Blum et al. 2006; Heyer et al. 2015). We estimated a common index of tree imbalance (Colless index) and calculated whether the mtDNA and NRY trees were more unbalanced than 1000 simulated trees generated under a Yule process (Bortolussi et al. 2006) (i.e. a simple pure birth process that assumes that the birth rate of new lineages is the same along the tree). We found that the NRY tree is more unbalanced than predicted by the Yule model (p-value=0.001), whereas the mtDNA tree is not significantly different from trees generated by the Yule model (p-value=0.628). It has been suggested that highly mobile hunter-gatherer societies, such as those typical of most of human prehistory, were polygynous bands (Dupanloup et al. 2003); similarly, nomadic horticulturalist Amazonian societies exhibit strong differences in reproductive success due to the common practice of polygyny, especially among community chiefs, whose offspring also enjoy a high fertility (Neel 1970; 1980; Neel and Weiss 1975).
Furthermore, a more recent expansion can be observed in the BSP based on the NRY, but not in the mtDNA BSP (Figure 5), indicating an expansion specifically in the paternal line. The reasons behind this recent male-biased population expansion, which starts ~3.5 kya, are as yet unclear. However, similar male-biased expansions have been observed in other studies using high-resolution NRY sequences (Batini et al. 2017; Karmin et al. 2015).
Related:
- Post-Neolithic Y-chromosome bottleneck explained by cultural hitchhiking and competition between patrilineal clans
- Ancient human parallel lineages within North America contributed to a coastal expansion
- Population structure in Argentina shows most European sources of South European origin
- Latin Americans show widespread Mediterranean and North African ancestry
- Patterns of genetic differentiation and the footprints of historical migrations in the Iberian Peninsula
- Ancient Patagonian genomes suggest origin and diversification of late maritime hunter-gatherers
- Ancient DNA reveals temporal population structure of pre-Incan and Incan periods in South‐Central Andes area
- Paternal lineages mainly from migrants, maternal lineages mainly from local populations in Argentina