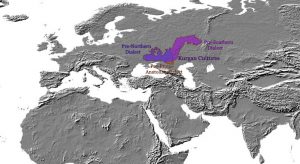
Map of Late Eneolithic migrations, from the collection of Maps of prehistoric migrations.
For context and reference, please read the book on Indo-European and Uralic migrations.
Click on each file to open or download the full version.
Current version: 6.3 (March 2021)
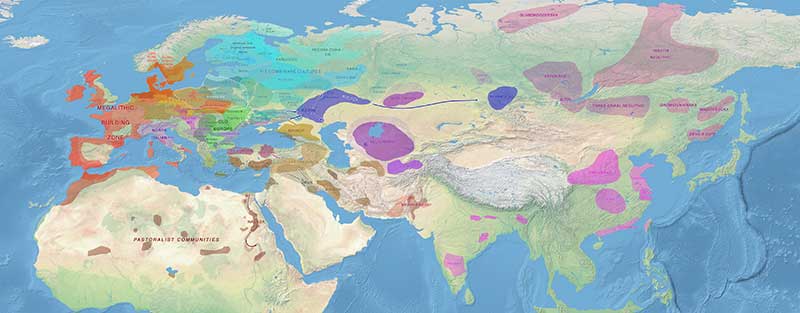
NOTE. As of September 2020, there is a new intermediate map showing the Final Eneolithic period, set between the previous Late Eneolithic and Early Chalcolithic maps.
Maps of Y-DNA, mtDNA, and ADMIXTURE
![]() Y-DNA Data (PDF). 28 March 2020.
Y-DNA Data (PDF). 28 March 2020.
![]() OpenLayers Y-DNA Map (slow to load!). 10 June 2020.
OpenLayers Y-DNA Map (slow to load!). 10 June 2020.
![]() mtDNA data (PDF). 28 March 2020.
mtDNA data (PDF). 28 March 2020.
![]() OpenLayers mtDNA Map (slow to load!). 3 June 2020.
OpenLayers mtDNA Map (slow to load!). 3 June 2020.
![]() ADMIXTURE K=7 (PDF). December 2019.
ADMIXTURE K=7 (PDF). December 2019.
Image with no labels (JPG). February 2019.
Older versions
Fifth version: 2019-2020
Fourth version: October 2017
Third version: September 2017
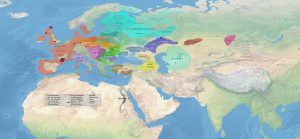
Second version: May 2017
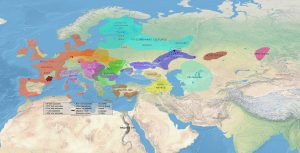
First version: February 2017
Older Maps: 2007