An open access article Complex patterns of admixture across the Indonesian archipelago, by Hudjashov et al. (2017), has appeared in Molecular Biology and Evolution, and clarifies further the Austronesian (AN) expansion.
Abstract:
Indonesia, an island nation as large as continental Europe, hosts a sizeable proportion of global human diversity, yet remains surprisingly under-characterized genetically. Here, we substantially expand on existing studies by reporting genome-scale data for nearly 500 individuals from 25 populations in Island Southeast Asia, New Guinea and Oceania, notably including previously unsampled islands across the Indonesian archipelago. We use high-resolution analyses of haplotype diversity to reveal fine detail of regional admixture patterns, with a particular focus on the Holocene. We find that recent population history within Indonesia is complex, and that populations from the Philippines made important genetic contributions in the early phases of the Austronesian expansion. Different, but interrelated processes, acted in the east and west. The Austronesian migration took several centuries to spread across the eastern part of the archipelago, where genetic admixture postdates the archeological signal. As with the Neolithic expansion further east in Oceania and in Europe, genetic mixing with local inhabitants in eastern Indonesia lagged behind the arrival of farming populations. In contrast, western Indonesia has a more complicated admixture history shaped by interactions with mainland Asian and Austronesian newcomers, which for some populations occurred more than once. Another layer of complexity in the west was introduced by genetic contact with maritime travelers from South Asia and strong demographic events in isolated local groups.
Among its results (emphasis is mine):
Most eastern Indonesian populations show traces of admixture that appear to reflect an expansion of AN speakers (Figure 4B, S3). There is a striking similarity between inferred events – each admixed population includes both a Philippine non-Kankanaey and western Indonesian-like source likely representing Holocene movements of Asian farming groups, as well as a Papuan-like source representing local indigenous ancestry. One reason for the lack of clear Taiwanese sources may be because the aboriginal populations of Taiwan were heavily affected by post-AN movements from mainland East Asia, most recently sinicization by Han Chinese, and thus no longer depict the ancestral AN gene pool (Mörseburg, et al. 2016). However, this notable pattern could equally be explained by the dominance of language and culture transfers during early phases of the Neolithic expansion from Taiwan into the Philippines, followed by people with predominantly Philippine ancestry driving later demic diffusion into the Indonesian archipelago. Interestingly, Mörseburg, et al. (2016), by using a different sample set and genotype-based analytical toolkit, indicated that the Kankanaey ethnic group from the Philippines is likely the closest living proxy of the source population that gave rise to the AN expansion. We did not detect this population among sources of admixture in eastern Indonesia, and therefore suggest that the place of individual Philippine groups in the AN expansion needs to be further addressed by better sampling in the Philippine archipelago.
Sumba and Flores, the two westernmost islands to the east of Wallace’s line, display a high proportion of Java and Bali surrogates in their AN admixing source. This suggests that the AN movement into eastern Indonesia, especially for Sumba and Flores, had earlier experienced some degree of genetic contact with western Indonesian groups. In contrast, the sources of AN admixture in Lembata, Alor, Pantar and Timor are dominated by Sulawesi (Figure 4B, S3, Table S3, S5). This generally agrees with expectations from the geography of the region, whereby AN groups exiting the southern Philippines were likely funneled into at least two streams, including a western path through Borneo and a central path through Sulawesi (Blust 2014).
Point estimates of genetic admixture times in eastern Indonesia lie within a narrow timeframe ranging between ca 185 BCE to 360 CE or 75 to 56 generations ago (95% CI 510 BCE – 475 CE or 87–52 generations) (Figure 4B, Table S3). These inferred dates are younger than some previous estimates (120–200 generations ago) (Xu, et al. 2012; Sanderson, et al. 2015; Sedghifar, et al. 2015). A major analysis of admixture in Indonesia estimated the date of AN contact in the eastern part of archipelago to be around 500 to 600 CE (ca 50 generations, CI estimates between 58–42 generations ago) (Lipson, et al. 2014), surprisingly young given the archaeological evidence. However, the study pooled a very small sample of genetically heterogeneous eastern Indonesian islands including, for example, Flores and Alor. As we show here (Figure 2, 4, 5, S3, Table S3, S5, S6), while the wave of AN speakers left a common genetic trace across the whole of eastern Indonesia, the details and dates of this contact vary considerably not only between islands (e.g., Flores and Alor), but also within individual islands (e.g., Flores Rampasasa vs. Flores Bama). The genetic dates, which were obtained here by denser geographical sampling of 8 eastern islands, a much larger number of individuals (28 per island on average) and a greater number of SNPs, are up to 30 generations older, predating the Common Era in many cases.
It therefore took migrants at least half a millennium to proceed from islands around Wallace’s line to the easternmost sampled part of eastern Indonesia. Nevertheless, observed dates for AN contact in eastern Indonesia are still approximately a millennium younger than the earliest Neolithic archaeological evidence in the region, and two explanations seem most likely here. First, the AN migration may have involved several waves of people leaving Taiwan, spanning multiple generations, which would bias date estimates later than the first arrival of the Neolithic archeological assemblage (Sedghifar, et al. 2015). Second, there may have been a substantial time gap between the spread of culture and technological traditions, and the beginning of extensive genetic contact between incoming farming groups and native inhabitants in Indonesia (Lansing, et al. 2011). The lack of considerable admixture with Papuan groups was recently noted in ancient Lapita individuals from Remote Oceania, whose genomes are mostly Asian and carry little to no Papuan ancestry, suggesting limited contact as they moved through Melanesia to previously uninhabited islands in the Pacific (Skoglund, et al. 2016). A lag in admixture between local and incoming Neolithic groups has also been observed in Europe, where hunter-gatherer and farming populations initially co-existed for nearly a thousand years without substantial genetic interaction (Malmström, et al. 2015).
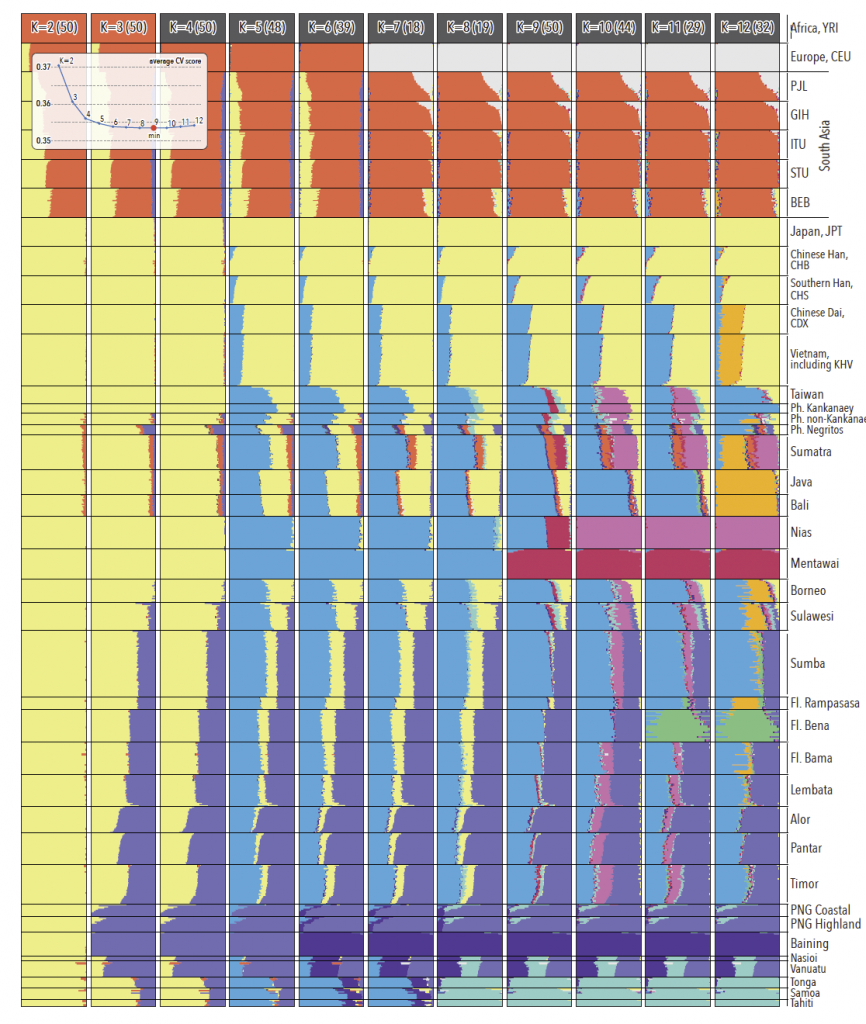 Ancestral genomic components in regional populations. For every K, the modal solution with the highest number of ADMIXTURE runs is shown; individual ancestry proportions were averaged across all runs from the same mode and the number of runs (out of 50) assigned to the presented solution is shown in parentheses. Average cross validation statistics were calculated across all runs from the same mode (insert). The minimum cross-validation score is observed at K=9. Note major ancestry components in Indonesia and ISEA – Papuan (light purple), mainland Asian (light yellow) and AN (light blue) – as well as major differences in the distribution of these three ancestries between eastern and western Indonesia. Populations from the Philippines and Flores are abbreviated as ‘Ph.’ and ‘Fl.’, respectively.
Ancestral genomic components in regional populations. For every K, the modal solution with the highest number of ADMIXTURE runs is shown; individual ancestry proportions were averaged across all runs from the same mode and the number of runs (out of 50) assigned to the presented solution is shown in parentheses. Average cross validation statistics were calculated across all runs from the same mode (insert). The minimum cross-validation score is observed at K=9. Note major ancestry components in Indonesia and ISEA – Papuan (light purple), mainland Asian (light yellow) and AN (light blue) – as well as major differences in the distribution of these three ancestries between eastern and western Indonesia. Populations from the Philippines and Flores are abbreviated as ‘Ph.’ and ‘Fl.’, respectively.
Featured images are taken from the article.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (https://creativecommons.org/licenses/by-nc/4.0/), which permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited. For commercial re-use, please contact [email protected]
Related posts: